Abstract
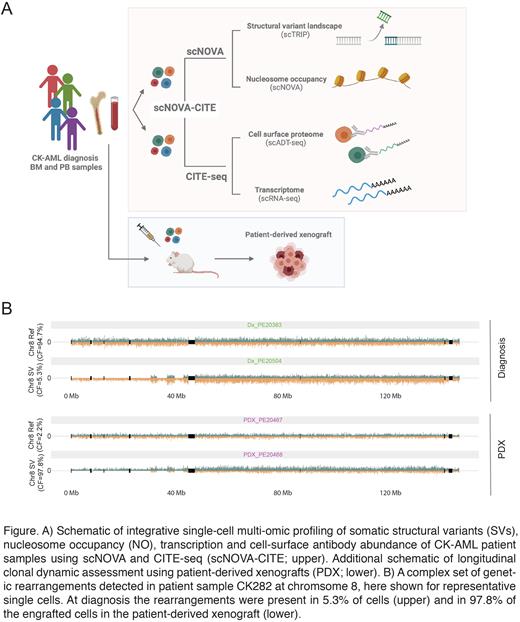
Genomic instability is a major driver of intra-tumor heterogeneity. While different single-cell technologies have emerged as powerful tools to study tumor heterogeneity, the complexity of somatic structural variants (SVs) in cancer and their connection to cell phenotype and functional outcome remain largely unexplored. To address this, we used Acute Myeloid Leukemia with complex karyotype (CK-AML) as a model disease entity to functionally dissect intra-patient heterogeneity on multiple levels. CK-AML is a heterogenous AML subgroup with dismal response to standard treatment. It remains understudied and poorly characterized at the molecular level, with mainstream techniques struggling to unravel the complexity of its SVs and to link them to molecular and clinical phenotypes. To improve our understanding of the molecular dynamics in CK-AML, we established an integrated single-cell multi-omics framework that combines the haplotype-resolved analysis of SVs and nucleosome occupancy (NO) profiling (scNOVA)(Jeong et al., 2021; Sanders et al., 2020) with concurrent immunophenotypic and transcriptomic profiling (CITE-seq)(Hao et al., 2021), here termed scNOVA-CITE (Figure A).
To investigate single-cell SV and NO landscapes in CK-AML, we generated single cell template strand-sequencing (Strand-seq)(Sanders, Falconer, Hills, Spierings, & Lansdorp, 2017) libraries from bone marrow or peripheral blood cells from four diagnostic CK-AML patient samples, and applied scNOVA(Jeong et al., 2021; Sanders et al., 2020) on 278 single-cell libraries. All samples carried multiple SVs, including somatic gains and losses of terminal chromosome regions, whole-chromosome aneuploidies, and a complex set of genetic rearrangements. We frequently detected extreme cases represented by a subset of cells with highly aberrant karyotypes that could be classified as hopeful monsters(Gerlinger et al., 2014; Markowetz, 2016). Using NO profiling, we assessed hierarchy composition by assigning each CK-AML cell to its corresponding normal hematopoietic differentiation state, and observed markedly varied abundances of different hematopoietic stem and progenitor cell (HSPC)-like states between the AMLs. Using CITE-seq, immunophenotypic and transcriptomic maps of 14,941 AML cells from the same CK-AML patient samples revealed strong transcriptional and immunophenotypic heterogeneity across all patients.
Next, we explored different layers of intra-patient heterogeneity by focusing on each leukemic sample individually. Two patients showed high genetic intra-patient heterogeneity and harbored multiple subclones at diagnosis with subclone-specific upregulation of genes associated with chemoresistance and adverse outcome. We followed the clonal dynamics of the subclones upon transplantation into sublethally irradiated NOD.Prkdcscid.Il2rgnull (NSG) mice, which revealed predominantly monoclonal engraftment and strongly reduced genomic instability in the respective patient-derived xenografts (Figure A-B). The engrafting clones could be traced back to a minor subclone in the original patient samples. Molecular analysis of the subclones in the diagnostic samples by scNOVA-CITE revealed converging characteristics leading to stem-like phenotypes. To exemplify the translational relevance of the clonal evolution analysis in mice, we analyzed cells from one of the patients during relapse and identified the engrafting clone to also drive relapse, recapitulating subclone-specific evolution during disease progression. Taken together, these data indicate that cytogenetic evolution resulting in stemness phenotype in CK-AML is advantageous for leukemic stem cell expansion in mice and drives disease progression in patients, unfolding novel ways to study regulation of stemness and to identify potential therapeutic targets.
By developing scNOVA-CITE, we established an integrative single-cell multi-omics framework allowing to characterize genetic, epigenetic, transcriptomic and cell surface proteomic properties of CK-AML patient cells. We provide unique insights into the genetic complexity of CK-AML, and evidence of the resulting functional outcomes.
Disclosures
Bullinger:Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees; Celgene/BMS: Honoraria, Membership on an entity's Board of Directors or advisory committees; Gilead: Honoraria, Membership on an entity's Board of Directors or advisory committees; Astellas: Honoraria; Bayer Oncology: Research Funding; Jazz Pharmaceuticals: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Abbvie: Honoraria, Membership on an entity's Board of Directors or advisory committees; Pfizer: Honoraria, Membership on an entity's Board of Directors or advisory committees; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.